Development and application of bioorthogonal chemical reactions on living systems, to provide innovative tools for life science research and develop innovative technologies for diagnostics and therapeutics through the chemical knowledge and tools.
1.Bioorthogonal chemistry and chemical tools development.
Development and application of bioorthogonal chemistry and chemical tools for life sciences.
Development and application of bioorthogonal chemistry and chemical tools for life sciences.
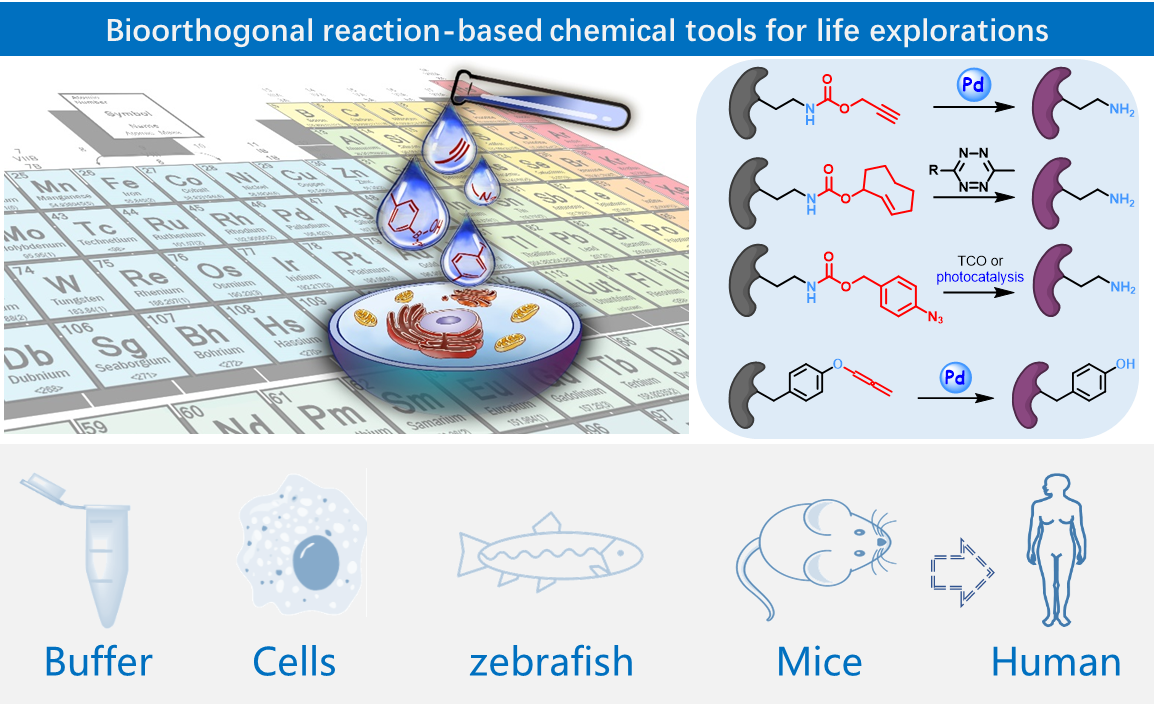
Recent representative work:
1), Spatiotemporally resolved dissection of subcellular proteome is crucial to our understanding of cellular functions in health and disease. We herein report a bioorthogonal and photocatalytic decaging-enabled proximity labeling strategy (CAT-Prox) for spatiotemporally resolved mitochondrial proteome profiling in living cells. Our systematic survey of the photocatalysts has led to the identification of Ir(ppy)2bpy as a bioorthogonal and mitochondria-targeting catalyst that allowed photocontrolled, rapid rescue of azidobenzyl-caged quinone methide as a highly reactive Michael acceptor for proximity-based protein labeling in mitochondria of live cells. Upon careful validation through in vitro labeling, mitochondria-targeting specificity, in situ catalytic activity as well as protein tagging, we applied CAT-Prox for mitochondria proteome profiling in living Hela cells as well as hard-to-transfect macrophage RAW264.7 cells with approximately 70% mitochondria specificity observed from up to 300 proteins enriched. Finally, CAT-Prox was further applied to the dynamic dissection of mitochondria proteome of macrophage cells upon lipopolysaccharide stimulation. By integrating photocatalytic decaging chemistry with proximity-based protein labeling, CAT-Prox offers a general, catalytic, and nongenetic alternative to the enzyme-based proximity labeling strategies for diverse live cell settings. (J. Am. Chem. Soc. 2021, 143, 18714-18720)
2), A universal gain-of-function approach for selective and temporal control of protein activity in living systems is crucial to understanding dynamic cellular processes. Here we report development of a computationally aided and genetically encoded proximal decaging (hereafter, CAGE-prox) strategy that enables time-resolved activation of a broad range of proteins in living cells and mice. Temporal blockage of protein activity was computationally designed and realized by genetic incorporation of a photo-caged amino acid in proximity to the functional site of the protein, which can be rapidly removed upon decaging, resulting in protein re-activation. We demonstrate the wide applicability of our method on diverse protein families, which enabled orthogonal tuning of cell signalling and immune responses, temporal profiling of proteolytic substrates upon caspase activation as well as the development of protein-based pro-drug therapy. We envision that CAGE-prox will open opportunities for the gain-of-function study of proteins and dynamic biological processes with high precision and temporal resolution.(Nature 2019, 569, 509-513)
Representative publications:
1, Ngai, W. S. C.; Yang, S.; Zeng, X.; Liu, Y.; Lin, F.; Wang, X.; Zhang, H.; Fan, X.; Chen, P. R., Bioorthogonally Activatable Base Editing for On-Demand Pyroptosis. J. Am. Chem. Soc. 2022. DOI: 10.1021/jacs.1c12924.
2, Huang, Z.; Liu, Z.; Xie, X.; Zeng, R.; Chen, Z.; Kong, L.; Fan, X.; Chen, P. R., Bioorthogonal Photocatalytic Decaging-Enabled Mitochondrial Proteomics. J. Am. Chem. Soc. 2021, 143 (44), 18714-18720.
3, Wang, J.; Wang, X.; Fan, X.; Chen, P. R., Unleashing the Power of Bond Cleavage Chemistry in Living Systems. ACS Cent. Sci. 2021, 7 (6), 929-943.
4, Liu, Y.; Zeng, R.; Wang, R.; Weng, Y.; Wang, R.; Zou, P.; Chen, P. R., Spatiotemporally resolved subcellular phosphoproteomics. Proc. Natl. Acad. Sci. USA. 2021, 118 (25), e2025299118.
5, Liu, S.; Lin, C.; Xu, Y.; Luo, H.; Peng, L.; Zeng, X.; Zheng, H.; Chen, P. R.; Zou, P., A far-red hybrid voltage indicator enabled by bioorthogonal engineering of rhodopsin on live neurons. Nat. Chem. 2021, 13 (5), 472-479.
6, Wang, J.; Liu, Y.; Liu, Y.; Zheng, S.; Wang, X.; Zhao, J.; Yang, F.; Zhang, G.; Wang, C.; Chen, P. R., Time-resolved protein activation by proximal decaging in living systems. Nature 2019, 569 (7757), 509-513.
7, Wang, X.; Liu, Y.; Fan, X.; Wang, J.; Ngai, W. S. C.; Zhang, H.; Li, J.; Zhang, G.; Lin, J.; Chen, P. R., Copper-Triggered Bioorthogonal Cleavage Reactions for Reversible Protein and Cell Surface Modifications. J. Am. Chem. Soc. 2019, 141 (43), 17133-17141.
8, Yao, Q.; Lin, F.; Fan, X.; Wang, Y.; Liu, Y.; Liu, Z.; Jiang, X.; Chen, P. R.; Gao, Y., Synergistic enzymatic and bioorthogonal reactions for selective prodrug activation in living systems. Nat. Commun. 2018, 9 (1), 5032.
9, Fan, X. Y.; Li, J.; Chen, P. R., Bioorthogonal chemistry in living animals. Natl. Sci. Rev. 2017, 4 (3), 300-302.
10, Li, J.; Chen, P. R., Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol. 2016, 12 (3), 129-37.
2, Huang, Z.; Liu, Z.; Xie, X.; Zeng, R.; Chen, Z.; Kong, L.; Fan, X.; Chen, P. R., Bioorthogonal Photocatalytic Decaging-Enabled Mitochondrial Proteomics. J. Am. Chem. Soc. 2021, 143 (44), 18714-18720.
3, Wang, J.; Wang, X.; Fan, X.; Chen, P. R., Unleashing the Power of Bond Cleavage Chemistry in Living Systems. ACS Cent. Sci. 2021, 7 (6), 929-943.
4, Liu, Y.; Zeng, R.; Wang, R.; Weng, Y.; Wang, R.; Zou, P.; Chen, P. R., Spatiotemporally resolved subcellular phosphoproteomics. Proc. Natl. Acad. Sci. USA. 2021, 118 (25), e2025299118.
5, Liu, S.; Lin, C.; Xu, Y.; Luo, H.; Peng, L.; Zeng, X.; Zheng, H.; Chen, P. R.; Zou, P., A far-red hybrid voltage indicator enabled by bioorthogonal engineering of rhodopsin on live neurons. Nat. Chem. 2021, 13 (5), 472-479.
6, Wang, J.; Liu, Y.; Liu, Y.; Zheng, S.; Wang, X.; Zhao, J.; Yang, F.; Zhang, G.; Wang, C.; Chen, P. R., Time-resolved protein activation by proximal decaging in living systems. Nature 2019, 569 (7757), 509-513.
7, Wang, X.; Liu, Y.; Fan, X.; Wang, J.; Ngai, W. S. C.; Zhang, H.; Li, J.; Zhang, G.; Lin, J.; Chen, P. R., Copper-Triggered Bioorthogonal Cleavage Reactions for Reversible Protein and Cell Surface Modifications. J. Am. Chem. Soc. 2019, 141 (43), 17133-17141.
8, Yao, Q.; Lin, F.; Fan, X.; Wang, Y.; Liu, Y.; Liu, Z.; Jiang, X.; Chen, P. R.; Gao, Y., Synergistic enzymatic and bioorthogonal reactions for selective prodrug activation in living systems. Nat. Commun. 2018, 9 (1), 5032.
9, Fan, X. Y.; Li, J.; Chen, P. R., Bioorthogonal chemistry in living animals. Natl. Sci. Rev. 2017, 4 (3), 300-302.
10, Li, J.; Chen, P. R., Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat. Chem. Biol. 2016, 12 (3), 129-37.
2.Protein engineering and omics.
Protein engineering and multi-omics technology.
Protein engineering and multi-omics technology.
Protein-protein interactions are of great significance for the regulation of cellular physiological activities. However, once a protein is isolated and purified, its original biological function is often lost and the corresponding protein-protein interactions and their functions are not able to be studied in vitro. Therefore, it is of great advantage to develop labeling techniques for protein interactions applicable to living systems. We have developed several highly specific protein photocrosslinking probes by combining genetic code expansion technology with photo crosslinking probes. The system is currently one of the most efficient and specific protein interactions capture tools available, and has been acquired by laboratories at nearly half a dozen universities or research institutions worldwide for the study of widespread protein-protein interactions in cells. We are currently focusing on the use of this technology to study histone dynamics of chemically modified interacting proteins and expect to use multidimensional histological techniques to resolve the molecular mechanisms by which nucleosomes, the basic unit of cellular chromatin, are regulated by chemical modifications.
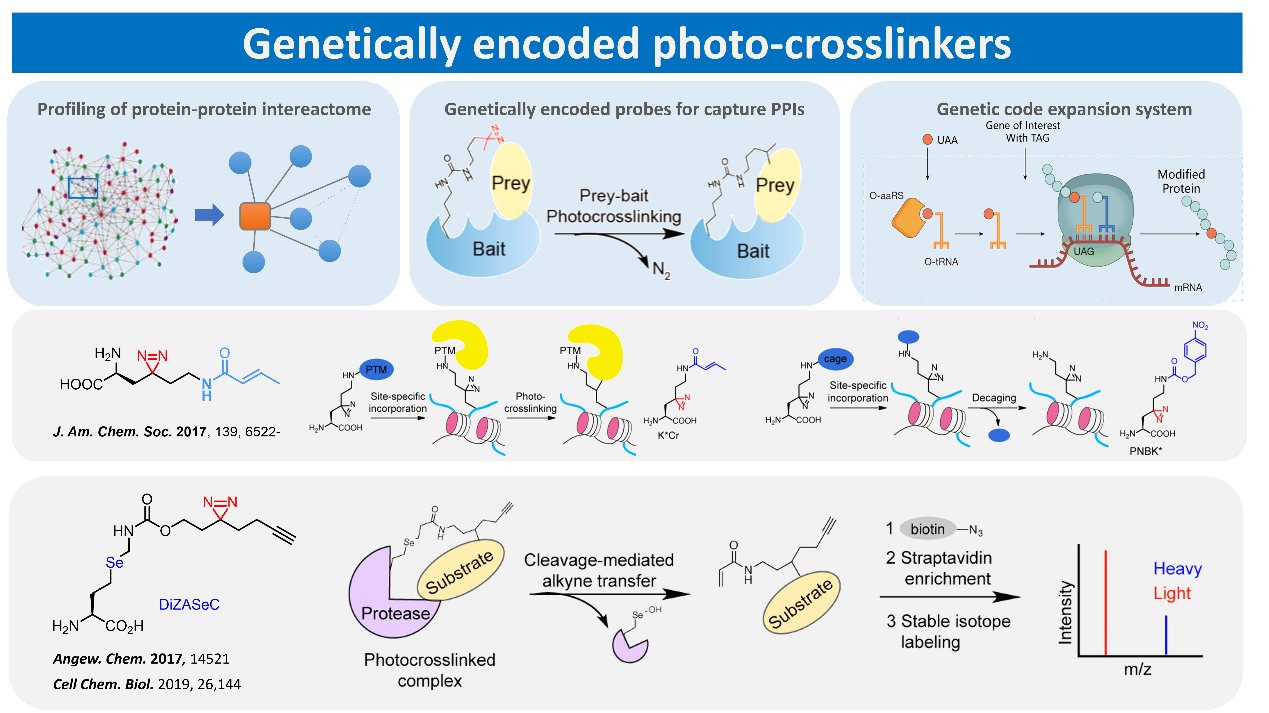
Representative publications:
1, Song H, Wang Y, Wang R, Zhang X, Liu Y, Jia G, Chen P. “SFPQ Is an FTO-Binding Protein that Facilitates the Demethylation Substrate Preference”, Cell Chem. Biol. (2020), 27, 283-91.
2, He D, Xie X, Yang F, Zhang H, Su H, Ge Y, Song H, Chen P. “Quantitative and Comparative Profiling of Protease Substrates through a Genetically Encoded Multifunctional Photocrosslinker”, Angew. Chem. Int. Ed. (2017), 56, 14521-5.
3, Xie X, Li X, Qin F, Lin J, Zhang G, Zhao J, Bao X, Zhu R, Song H, Li X, Chen P. “Genetically encoded photoaffinity histone marks”, J. Am. Chem. Soc. (2017), 139, 6522-5.
4, Yang Y, Song H, He D, Zhang S, Dai S, Xie X, Lin S, Hao Z, Zheng H, Chen P, “Genetically encoded releasable photocrosslinking strategies for studying protein-protein interactions in living cells”, Nat. Protocol. (2017), 12, 2147-68.
5, Zhang S, He D, Yang Y, Lin S, Zhang M, Dai S, Chen P. “Comparative proteomics reveal distinct chaperone-client interactions in supporting bacterial acid resistance”, Proc. Natl. Acad. Sci. USA. (2016), 113, 10872-7.
2, He D, Xie X, Yang F, Zhang H, Su H, Ge Y, Song H, Chen P. “Quantitative and Comparative Profiling of Protease Substrates through a Genetically Encoded Multifunctional Photocrosslinker”, Angew. Chem. Int. Ed. (2017), 56, 14521-5.
3, Xie X, Li X, Qin F, Lin J, Zhang G, Zhao J, Bao X, Zhu R, Song H, Li X, Chen P. “Genetically encoded photoaffinity histone marks”, J. Am. Chem. Soc. (2017), 139, 6522-5.
4, Yang Y, Song H, He D, Zhang S, Dai S, Xie X, Lin S, Hao Z, Zheng H, Chen P, “Genetically encoded releasable photocrosslinking strategies for studying protein-protein interactions in living cells”, Nat. Protocol. (2017), 12, 2147-68.
5, Zhang S, He D, Yang Y, Lin S, Zhang M, Dai S, Chen P. “Comparative proteomics reveal distinct chaperone-client interactions in supporting bacterial acid resistance”, Proc. Natl. Acad. Sci. USA. (2016), 113, 10872-7.
3.Immune chemical biology.
Interactions between biomacromolecules as well as cells in the regulations and interventions of immune systems.
Cancer immunotherapy is one of the most promising frontier research directions, and in situ resolution of tumor microenvironment is an important foundation for tumor immunotherapy. We have developed chemical technologies to achieve in situ real-time resolution of tumor microenvironment including cell-cell interaction capture.
Tumor microenvironment dissection: Interactions and information transfer between different cells play key roles in numerous life activities including immune response, organ development, and neurotransmission. Currently, most methods to study cellular interactions require known cell types to be involved, making it difficult to discover or study unknown cellular interactions in complex living environments. To address this challenge, Peng Chen's group has achieved in situ capture of dynamic interactions between cells with the help of "directed evolution" technology and "proximity labeling" strategy. This technique, named EXCELL (Enzyme-mediated proximal cell labeling), provides a powerful tool for studying cell-cell interactions by displaying the sorting enzyme (mgSrtA) that the authors have evolved in a targeted manner on the surface of the cells to be studied, allowing in situ capture and identification of the cells interacting with them.(J. Am. Chem. Soc. 2019, 141, 5, 1833–1837)
Targeted degradation of membrane proteins: Membrane proteins are key players in cellular interactions and cell signaling, differentiation and proliferation, stress response and other life processes. They directly influence the development of many diseases including malignant tumors and neuropathic pain, and are an important class of drug targets. However, conventional drugs act by blocking the interaction of the extracellular region of membrane proteins with ligands or inhibiting the activity of their intracellular regions, which often suffer from incomplete inhibition or induce drug resistance. To address this problem, our team developed a mass spectrometry-based covalent single-domain antibody (GlueBody) screening method and obtained GlueBody targeting PD-L1, which led to the development of a universal membrane protein degradation technology, GlueTAC, which provides a powerful tool for eliminating abnormal membrane proteins and can overcome the problem of drug resistance due to long-term drug use (e.g., EGFR-targeted inhibitors in lung cancer therapy). This technology provides a powerful tool for eliminating abnormal membrane proteins and overcoming resistance problems associated with long-term drug use (e.g., EGFR-targeted inhibitors in lung cancer therapy), opening up an important direction for the development of antibody drugs.(J. Am. Chem. Soc. 2021, 143 (40), 16377-16382)
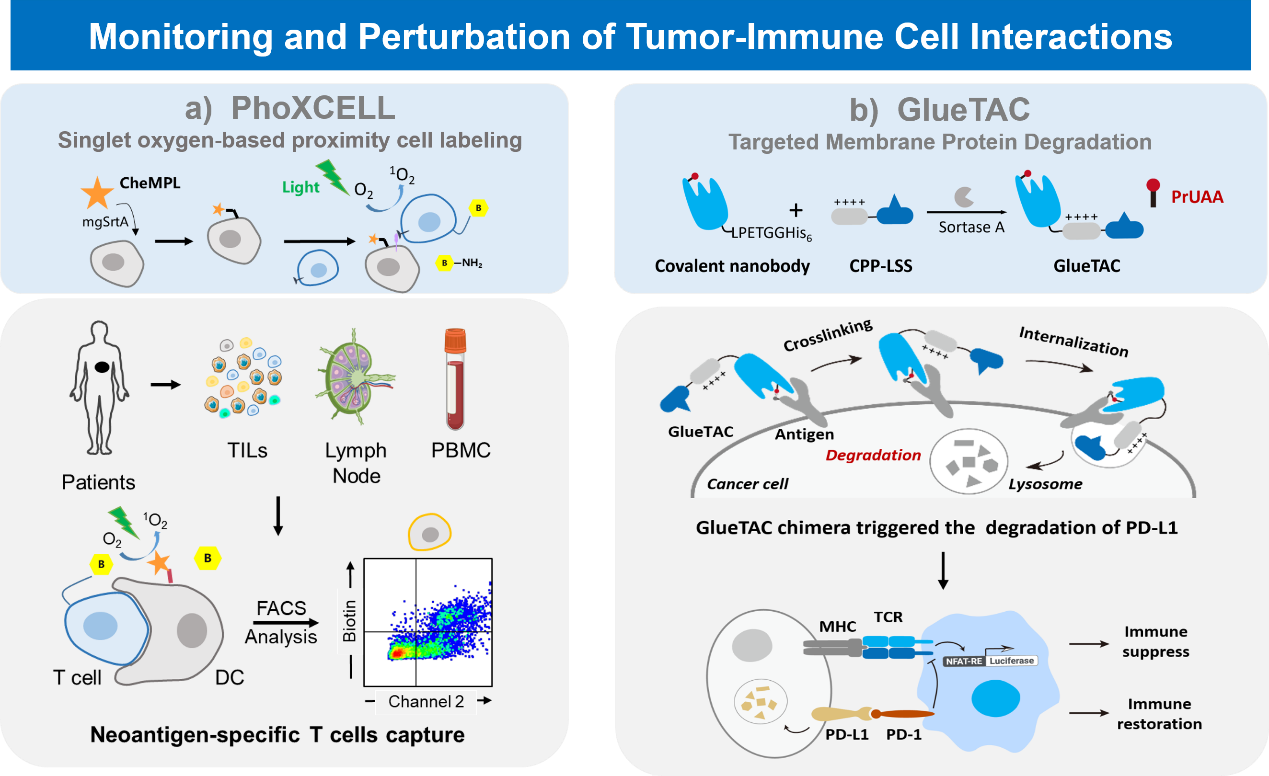
Representative publications:
1, Liu, H.; Luo, H.; Xue, Q.; Qin, S.; Qiu, S.; Liu, S.; Lin, J.; Li, J. P.; Chen, P. R., Antigen-Specific T Cell Detection via Photocatalytic Proximity Cell Labeling (PhoXCELL). J. Am. Chem. Soc. 2022, DOI: 10.1021/jacs.2c00159.
2, Ge, Y.; Chen, L.; Liu, S.; Zhao, J.; Zhang, H.; Chen, P. R., Enzyme-Mediated Intercellular Proximity Labeling for Detecting Cell–Cell Interactions. J. Am. Chem. Soc. 2019, 141 (5), 1833-1837.
3. Zhang, H.; Han, Y.; Yang, Y.; Lin, F.; Li, K.; Kong, L.; Liu, H.; Dang, Y.; Lin, J.; Chen, P. R., Covalently Engineered Nanobody Chimeras for Targeted Membrane Protein Degradation. J. Am. Chem. Soc. 2021, 143 (40), 16377-16382..
2, Ge, Y.; Chen, L.; Liu, S.; Zhao, J.; Zhang, H.; Chen, P. R., Enzyme-Mediated Intercellular Proximity Labeling for Detecting Cell–Cell Interactions. J. Am. Chem. Soc. 2019, 141 (5), 1833-1837.
3. Zhang, H.; Han, Y.; Yang, Y.; Lin, F.; Li, K.; Kong, L.; Liu, H.; Dang, Y.; Lin, J.; Chen, P. R., Covalently Engineered Nanobody Chimeras for Targeted Membrane Protein Degradation. J. Am. Chem. Soc. 2021, 143 (40), 16377-16382..

