As the workhorse of life, proteins offer exceptional promise as next-generation biomaterials due to their precisely programmable structures, diverse bioactivities, and intrinsic biodegradability. However, achieving high mechanical strength, dynamic responsiveness, and biological activity simultaneously in fully protein-based systems remains challenging. Traditional protein hydrogels rely primarily on either chemical crosslinking (robust yet irreversible) or physical interactions (weak but reversible), limiting their ability to balance stability and tunability. Inspired by topological engineering strategies in synthetic polymers—such as entanglement and interlocked rings—Prof. Wen-Bin Zhang’s team sought to integrate mechanical reinforcement into protein materials without compromising biofunctionality.
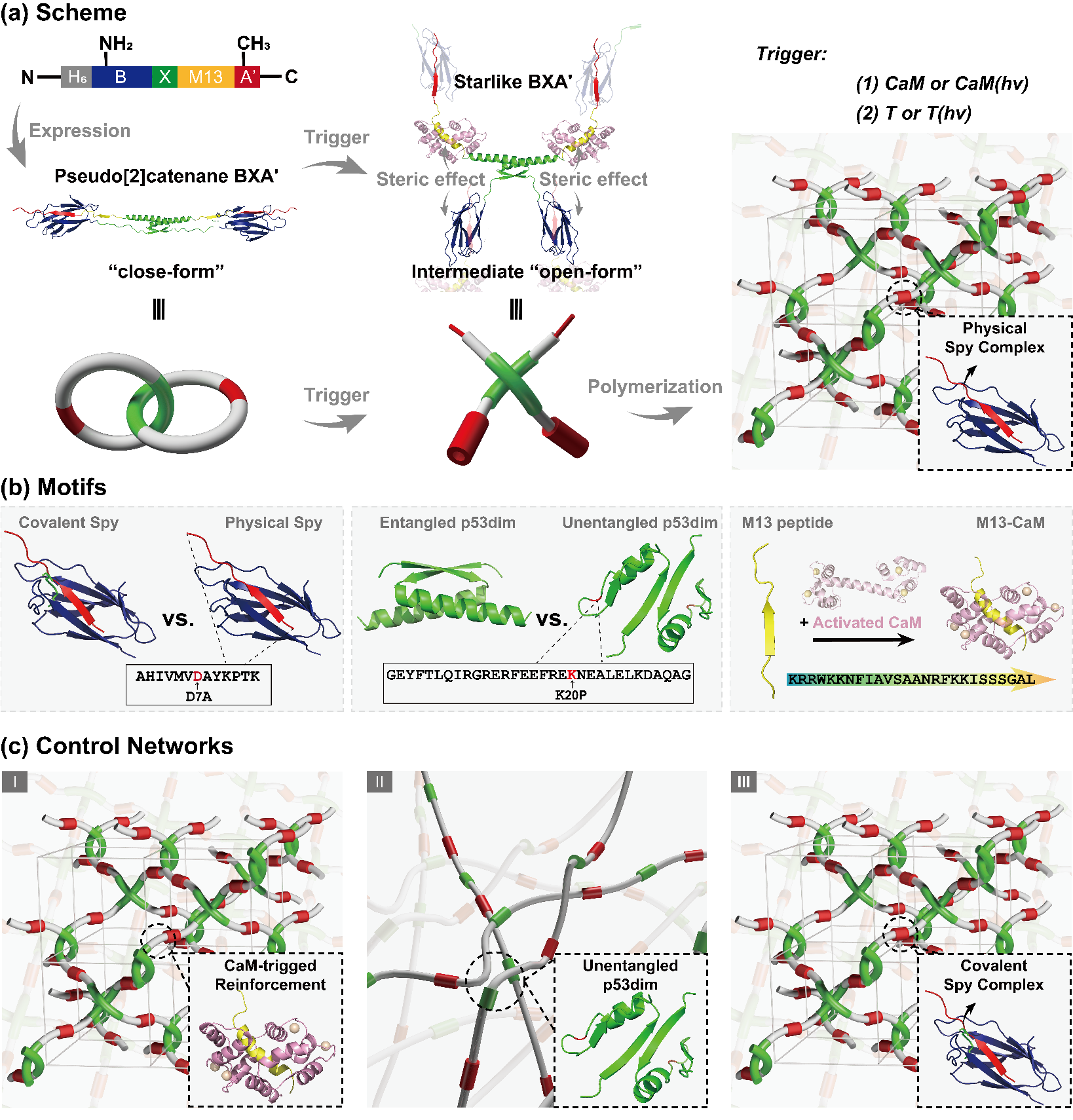
Figure 1. Design of interwoven protein network with topologically confined micro-association
The study reports an all-protein interwoven network built from a pseudo[2]rotaxane-like building block (Figure 1), BXA′, which combines three modular elements: a p53dim domain that enables topological entanglement, a SpyTag(DA)-SpyCatcher pair providing dynamic association, and a CaM–M13 module serving as a responsive trigger. Upon increasing concentration or applying external stimuli such as CaM binding or UV illumination, BXA′ transitions from a closed to a star-shaped conformation, forming physically crosslinked interwoven networks.
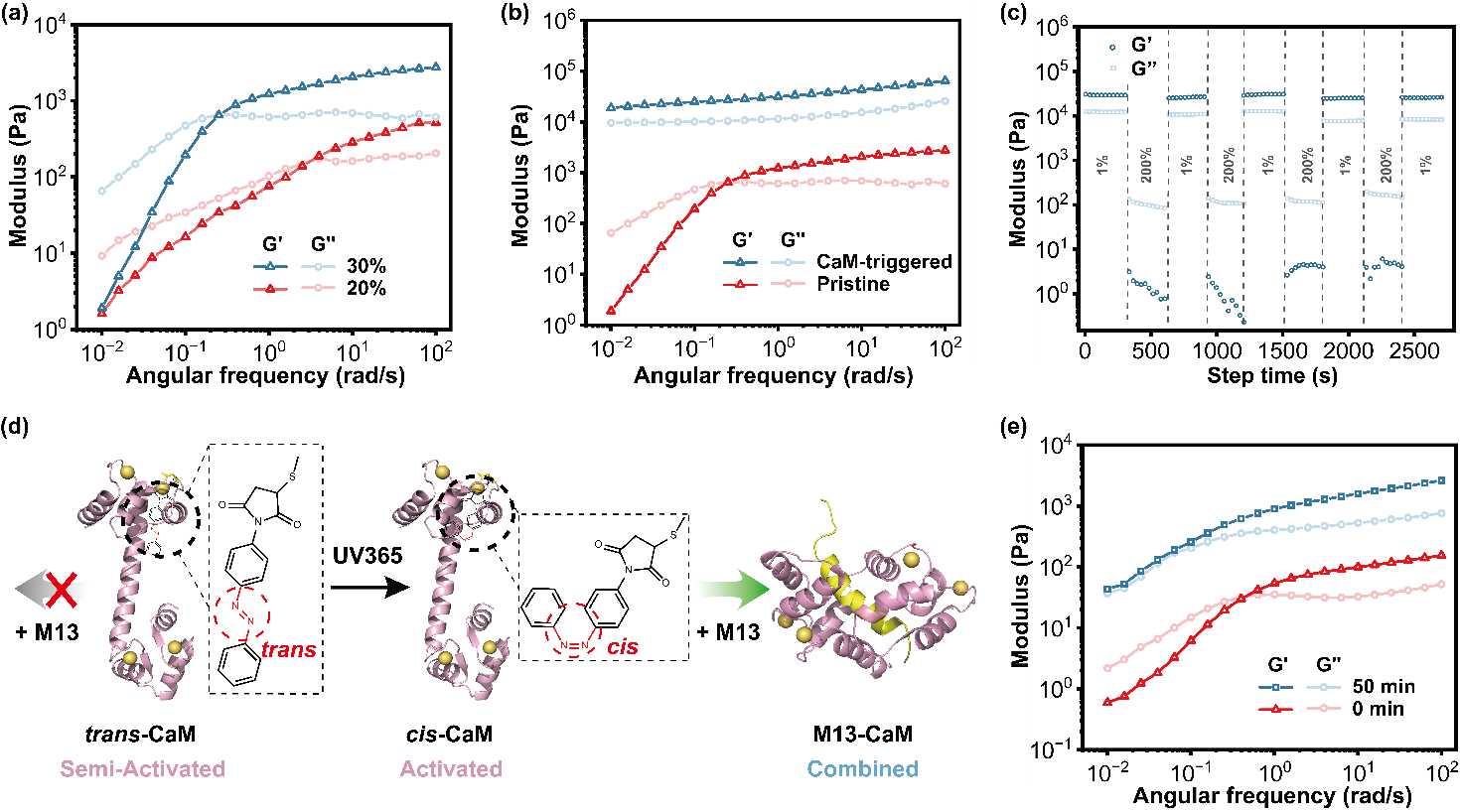
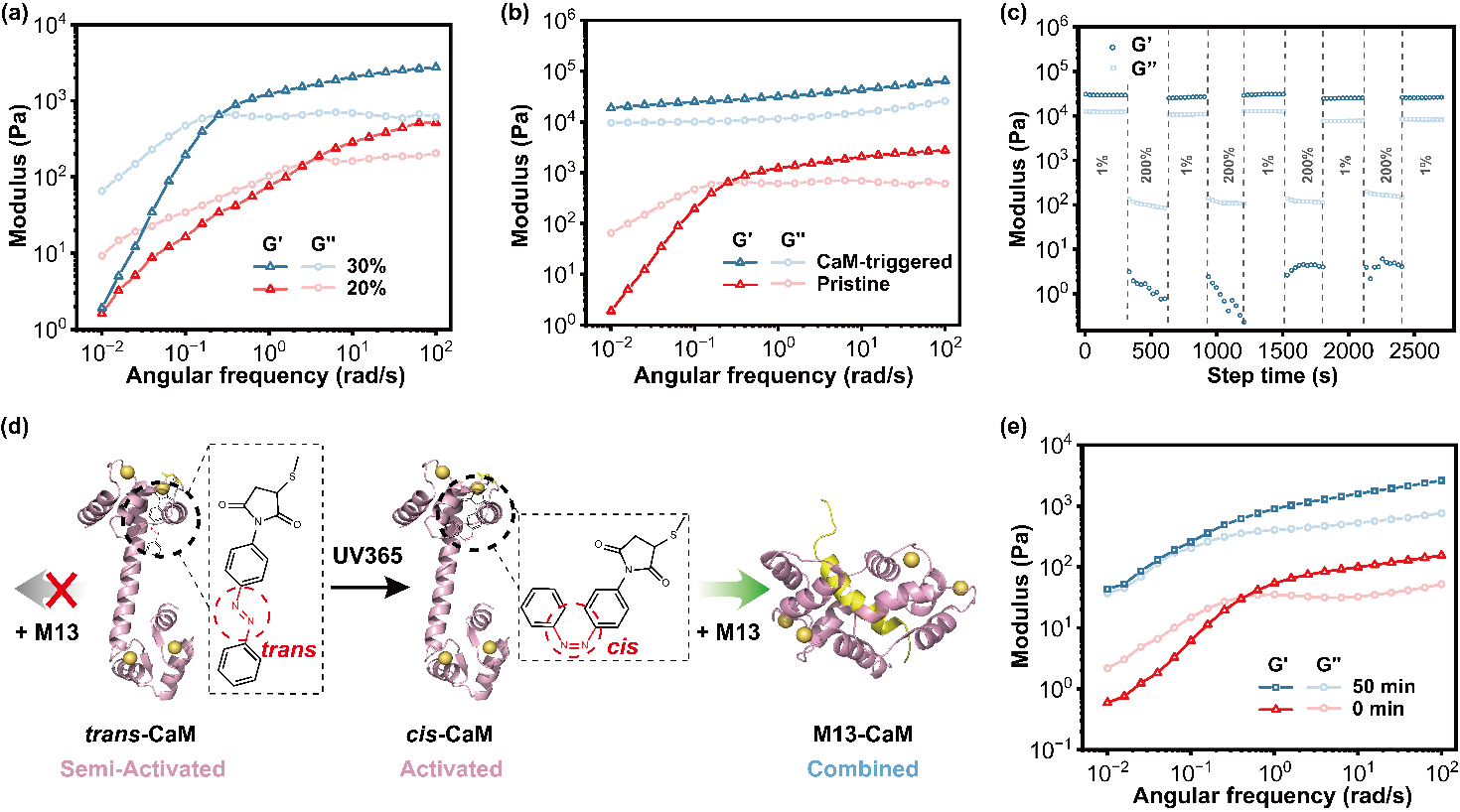
Figure 2. Mechanical properties of pristine and CaM-triggered BXA′ networks.
Under CaM-triggered conditions, the storage modulus (G′) of the BXA′ network increases with concentration, reaching over 60 kPa at 30 wt% (Figure 2). Subsequent thermal treatment (~70 °C) or near-infrared photothermal activation induces topologically confined micro-phase separation, boosting G′ to approximately 100 kPa. Atomic force microscopy measurements revealed a Young’s modulus approaching 1 MPa, positioning this material among the strongest known fully protein-based physical hydrogels. Moreover, the network exhibits remarkable self-healing and multi-stimuli responsive properties, achieving a unified balance of high strength and dynamic adaptability.
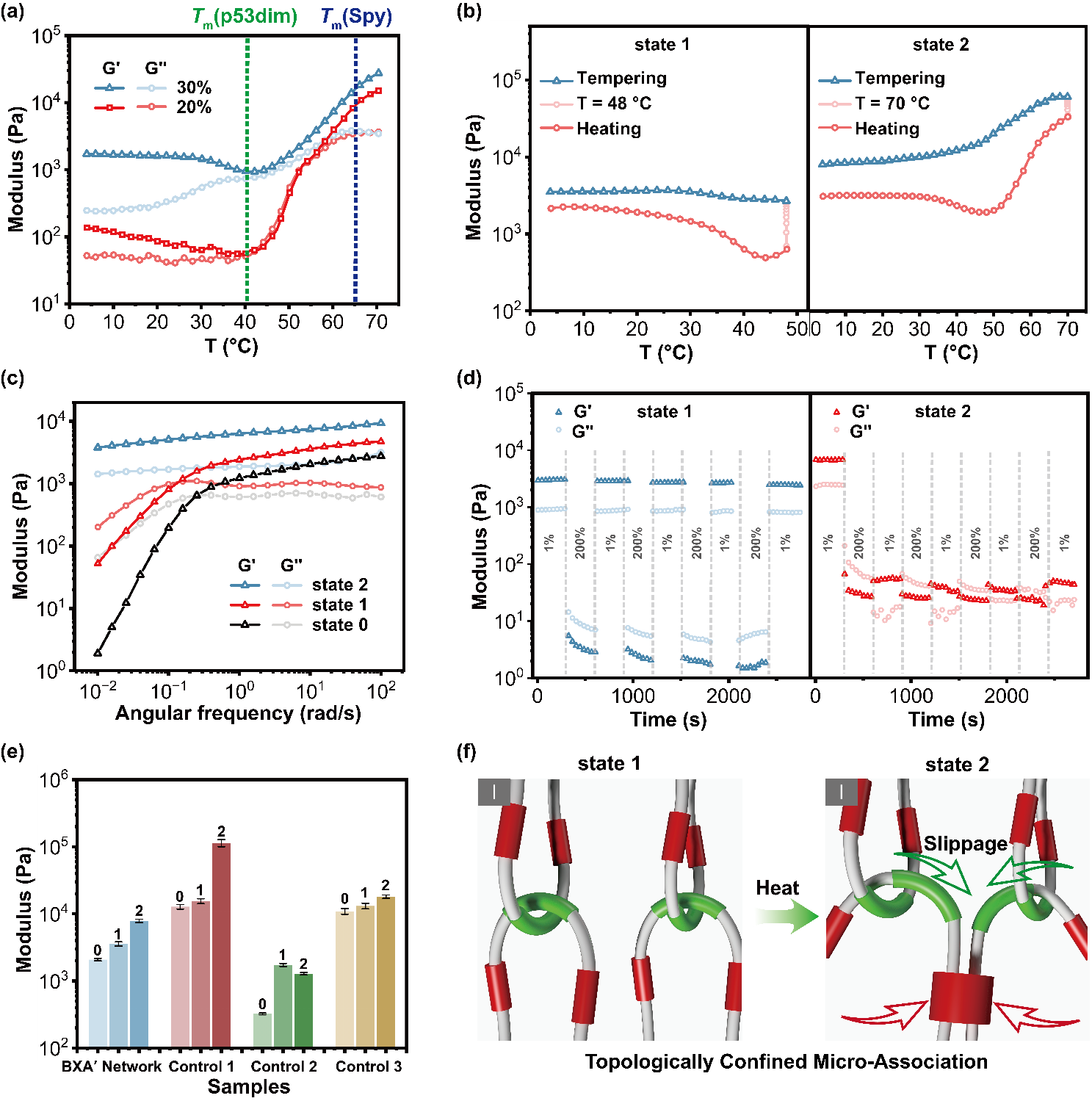
Figure 3. Mechanical behavior and mechanism of interwoven protein networks with topologically confined micro-association.
Mechanistic studies show that topological confinement within the interwoven network restricts the unfolding and aggregation of the SpyTag–SpyCatcher complex at elevated temperatures, preventing macroscopic phase separation while maintaining structural integrity (Figure 3). Compared to untangled or covalently crosslinked controls, the interwoven network demonstrated superior mechanical robustness, thermal stability, and erosion-resistant properties.
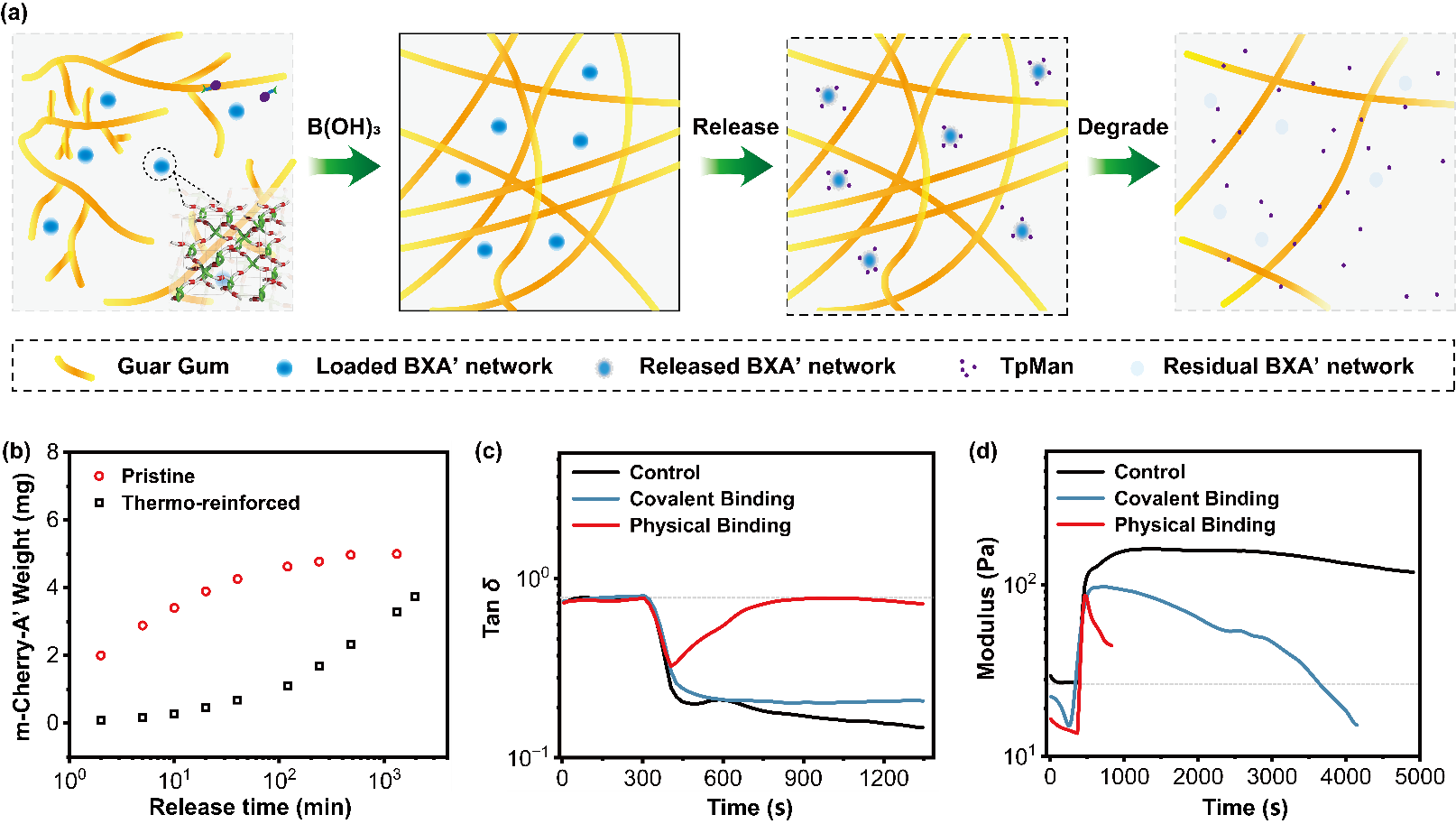
Figure 4. Thermally reinforced BXA′ network immobilizing TpMan for controlled guar-gum degradation.
Furthermore, the team explored the use of this enhanced network for enzyme immobilization and controlled release of thermophilic endo-mannanase (TpMan, Figure 4). By modulating covalent and non-covalent interactions between the enzyme and the network, they achieved tunable release kinetics and controlled guar gum degradation, highlighting its potential in time-programmed catalytic or industrial processes such as delayed gel breaking in oil recovery.
In summary, this work introduces a robust and versatile design strategy that integrates topological engineering principles into protein-based materials. The resulting networks combine unique topological architectures, outstanding mechanical performance, stimuli-responsiveness, and functional versatility. Beyond its implications for biomaterials design, this study provides further insights into the fundamental relationship between molecular topology, structure, and macroscopic properties—offering a sustainable approach for applications spanning biomedicine, catalysis, and smart materials.
The corresponding authors of this work are Prof. Wen-Bin Zhang, Dr. Wen-Hao Wu and Dr. Yajie Liu. Tingjie Xu, a PhD student, is the first author of the paper. Other contributors include Dr. Yibin Sun, Dr. Lianjie Xu, Yu-Xiang Wang, Fengyi Jiang, Bo Hou and Ziyi Meng. The work was supported by the National Key R&D Program of China, the Shenzhen Medical Research Fund, the National Natural Science Foundation of China, the Beijing National Center for Molecular Sciences, and the Boya Postdoctoral Fellowship of Peking University.
Original link for the paper: https://onlinelibrary.wiley.com/doi/10.1002/anie.202516010